Overview
 How does Mycobacterium tuberculosis adapt to stress and regulate its physiology over both short and long timescales to maximize survival in the harsh environment of the human host? Stress response is intimately linked to the regulation of gene expression. To learn how M. tuberculosis tolerates the challenges of infection, we need to understand the regulatory processes that control Mycobacterial stress response systems. We approach this problem in multiple ways, using both M. tuberculosis and its avirulent relative M. smegmatis.
How does Mycobacterium tuberculosis adapt to stress and regulate its physiology over both short and long timescales to maximize survival in the harsh environment of the human host? Stress response is intimately linked to the regulation of gene expression. To learn how M. tuberculosis tolerates the challenges of infection, we need to understand the regulatory processes that control Mycobacterial stress response systems. We approach this problem in multiple ways, using both M. tuberculosis and its avirulent relative M. smegmatis.
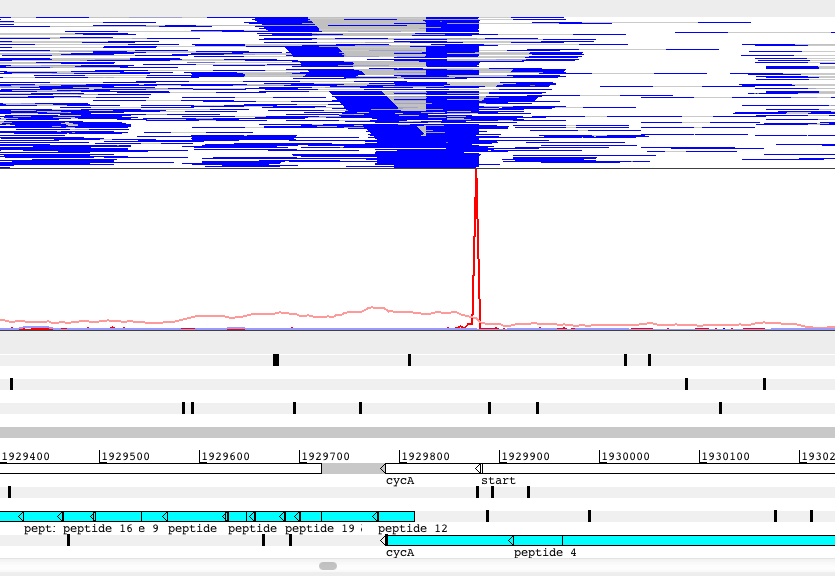
Regulation of transcript abundance is controlled by both transcription and RNA degradation. We use next-generation sequencing to map transcription start sites and RNA cleavage sites transcriptome-wide in order to understand the contributions of each of these processes to net changes in transcript abundance, revealing the mechanisms mycobacteria use to adapt to stressors such as hypoxia, oxidative stress, and antibiotics.
Our work is supported by NSF CAREER award 1652756, NIH/NIAID 1 P01 AI143575-01A1, and NIH/NIAID 1 R21 AI151481-01.
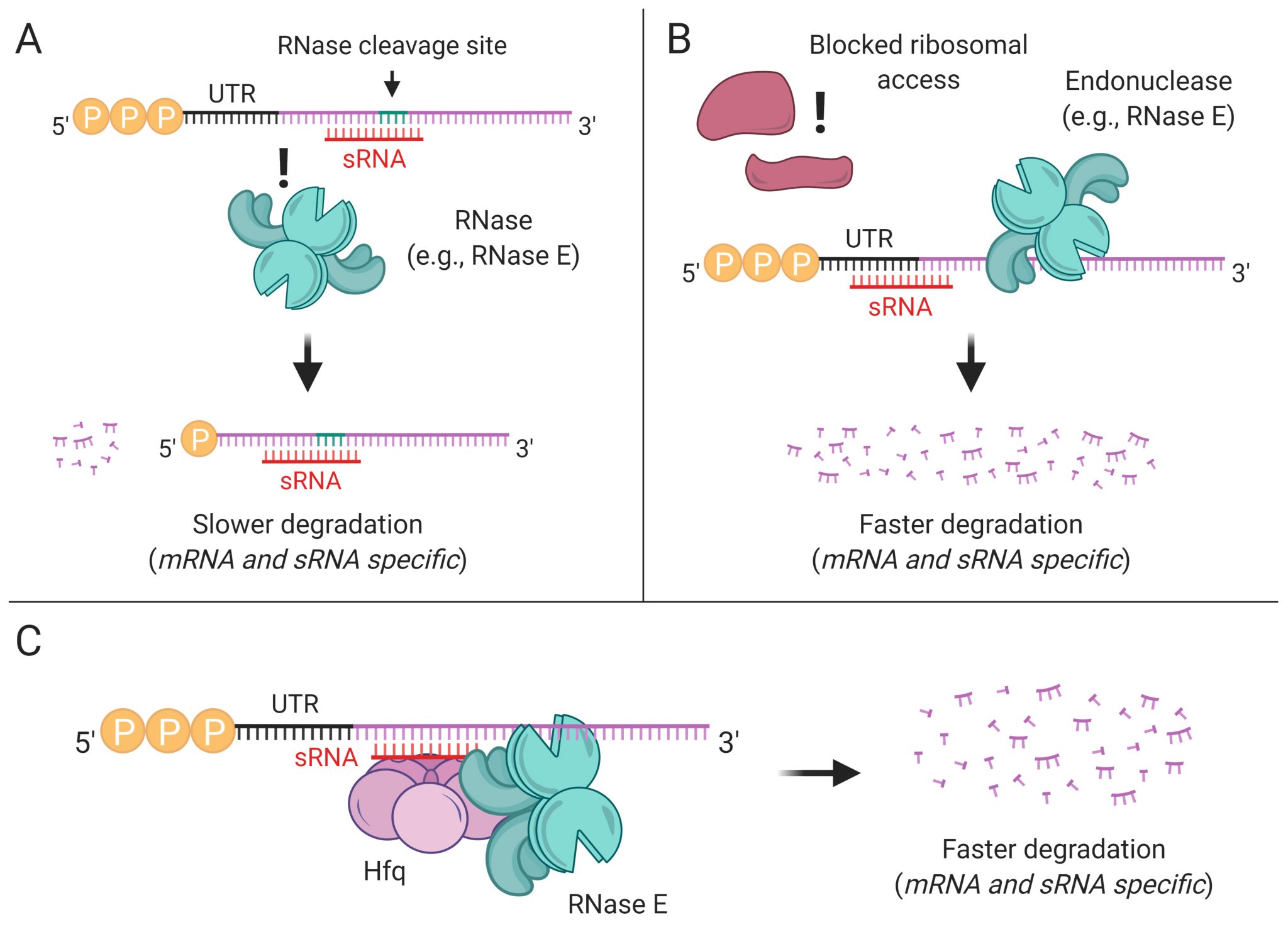
RNases
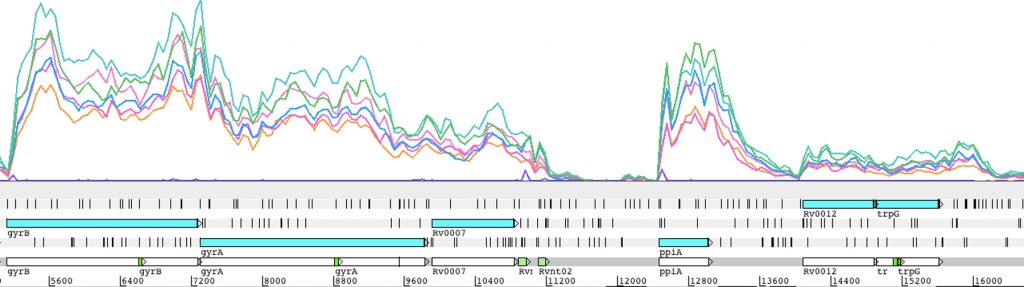
RNA processing and degradation are carried out by a suite of RNases whose roles and regulation in mycobacteria are largely unknown. We are genetically disrupting components of the RNA metabolism machinery and determining the consequences at the levels of both transcriptome and phenotype.
Transcript stability and stress tolerance
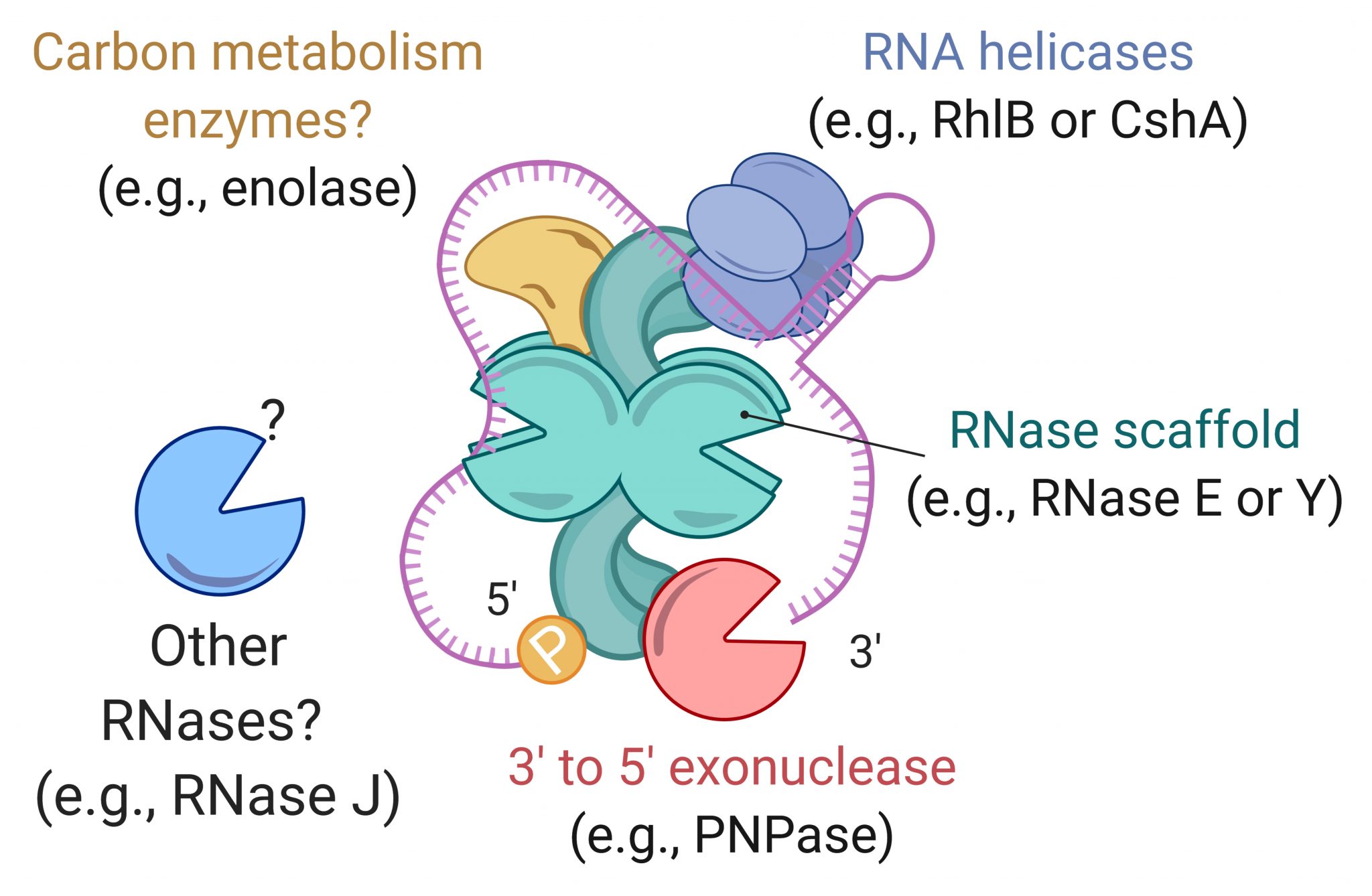
Transcript degradation rates are controlled by both gene-specific and global mechanisms. We are investigating the mechanistic basis of transcript degradation rate in mycobacteria in order to understand how regulation of transcript stability contributes to phenotypic adaptation to changing conditions.
Transcriptional regulation
We have found that DNA methylation and alternative sigma factors both contribute to the mycobacterial response to hypoxia, and are investigating the mechanisms involved.