About

Professor Reeta Rao is a leader in the field of molecular genetics and genomics and has affiliate appointments at the Broad Institute of MIT and Harvard (Cambridge) as well as the Institute of Drug Resistance at the Univ. of Mass Chan Medical School (Worcester). Her primary research activities are focused on emerging infectious diseases, specifically understanding, and managing fungal diseases. Students and research associates in her laboratory are trained to use a variety of high biochemical, molecular-genetic, and genomic tools to study host-microbe interactions to explore fungal virulence strategies and identify novel therapeutics in a high throughput fashion.
In addition, Prof. Rao is committed to the career and professional development of scientists at all levels of training. To keep researchers engaged in science, she has spree headed several workforce development opportunities aimed at recruiting, retaining, and improving the critical skills, knowledge, and resources required for academia as well as the industry. At the undergraduate level she is deeply engaged in the Global Health and Pre-Health programs. At the graduate level, she has championed several programs from micro credentials to certificates and MS degree programs to provide advanced coursework and laboratory techniques applicable to the biotechnology industry as well as upskilling for workforce development. She has served as the inaugural Associate Dean of Graduate studies.
Prof. Rao is a member of several professional societies and fellow of the American Academy of Microbiology (AAM fellow) and American Association for the Advancement of Science (AAAS fellow). She is a recipient of the Waksman outstanding Teacher award from the Society of Industrial Microbiology and Biotechnology.
Research
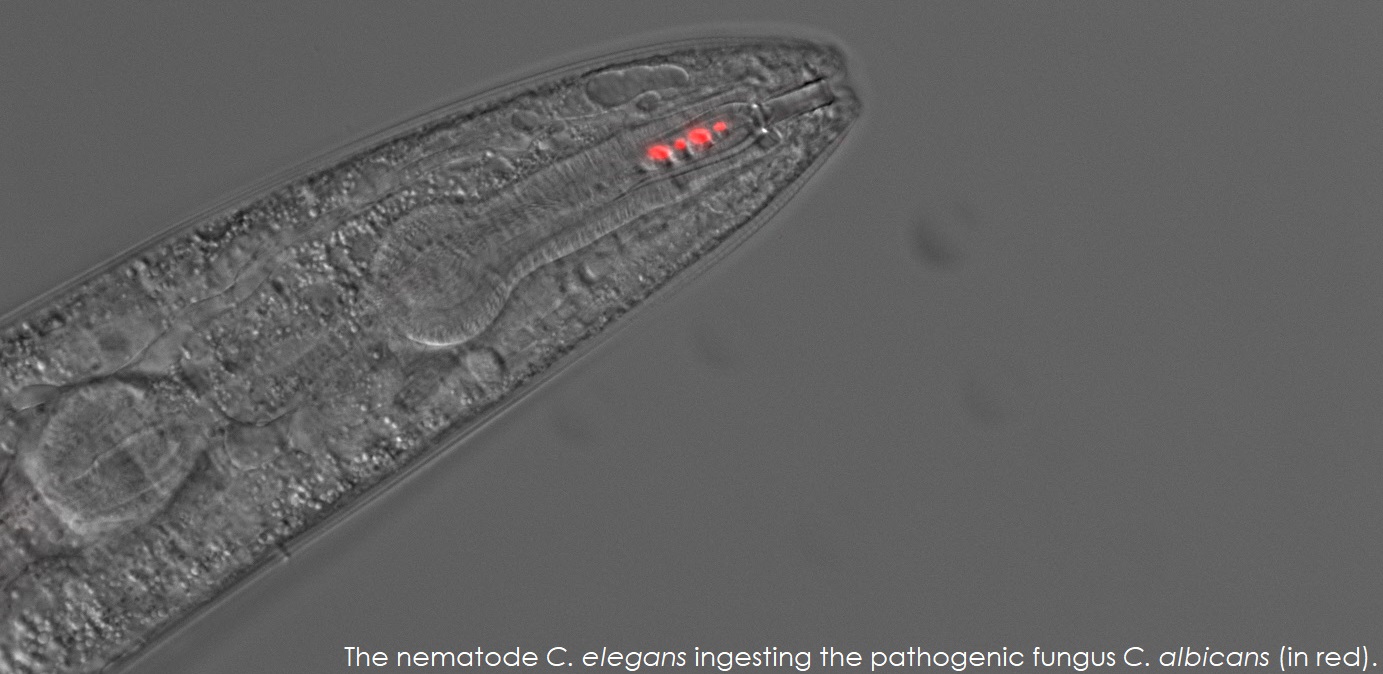
The goal of the Rao Lab is to understand and manage fungal infectious diseases. Our laboratory has been in the forefront of developing assays using Caenorhabditis elegans as a live animal model to study fungal infectious agents and microbial interactions in the gut.
Project 1 – Defining the Gut-Brain Axis
| We have discovered that pathogenic Fungi such as Candida albicans secrete metabolites that attract the nematode Caenorhabditis elegans via the activation of sensory neurons. C. elegans ingests the pathogen which colonizes the intestine and mounts an immune response. Later stages of disease manifests as intestinal distension which induces expression of the neuromodulator to produce neurotransmitters that allows the animal to learn to avoid the pathogen, C. albicans. This learned avoidance behavior is transmitted to the progeny via epigenetic mechanisms. | |
The goals of this project are to –
|
|
Project 2 – Probiotics, mechanism of action
| Probiotics are live microorganisms that are beneficial to health and wellbeing when consumed or applied to the body. They are thought to boost host immunity, prevent growth of pathogenic microbes, neutralize their toxins, and improve digestion. We have identified several novel probiotics that can be used to limit antimicrobial usage thereby reducing the chance emerging drug resistant microbes. | |
The goals of this project are to –
|
|
Project 3 – Antifungal Drug Discovery
| Emerging drug resistant microbes is a clear and present threat to global public health. Biofilm formation is a key strategy adopted by microbes to gain resistance. We have developed high throughput methods to screen for putative compounds that inhibit biofilm formation. Such compounds present alternative strategies to combat resistance of pathogens. | |
| The goal of this project is to identify compounds that inhibit microbial adhesion. | |
Publications

Upskilling the cell therapy manufacturing workforce: design, implementation, and evaluation of a massive open online course (Submitted)
Authors: Barone P, Duguid J, Keumurian F, Neufeld C, RAO R, Rolle M, Skrip B, Springs S, Vega H, Van Vliet K, Wolfrum J, Yang M
A correlative study of the genomic underpinnings of virulence traits and drug tolerance of Candida auris
Authors: YANG B., Vaisvil B, Schmitt D, COLLINS JH, Young, EM, Kapatral V, RAO RP
Source: bioRxiv doi: https://doi.org/10.1101/2023.04.07.536049
Drop-Seq: Massively Parallel Single-Cell RNA-Seq of Saccharomyces cerevisiae and Candida albicans
Authors: Dohn R., Xie B, Back R, Selewa A, Eckart H, RAO RP, Basu A
Source: Vaccines Dec 27;10(1):30
Genetic basis for probiotic yeast phenotypes revealed 2 by nanopore sequencing
Authors: COLLINS, J H., Kunyeit, L., Weintraub, S., Sharma, N., White, C., Haq, N., Anu-Appaiah, K.A., RAO, RP*. and Young, E. M* (* Co-corresponding authors)
Source: Accepted manuscript in G3.
Yeasts originating from fermented foods, their potential as probiotics and therapeutic implication for human health and disease
Authors: Kunyeit, L., RAO, R.P*. and Anu-Appaiah, K.A* (*Co-corresponding authors)
Source: Critical Reviews in Food Science and Nutrition, pp.1-12.
Secondary metabolites from food-derived yeasts inhibit virulence of Candida albicans
Authors: Kunyeit, L., Kurrey, N.K., Anu-Appaiah, K.A. and RAO, R.P.
Source: Mbio, 12(4), pp.e01891-21.
Application of probiotic yeasts on candida species associated infection
Authors: Kunyeit, L., KA, A.A. and RAO, R.P.
Source: Journal of Fungi, 6(4), p.189.
Using COVID-19 as a teaching tool in a time of remote learning: A workflow for bioinformatic approaches to identifying candidates for therapeutic and vaccine development
Authors: BRYCE S, HEATH KN, ISSI L, Ryder EF, RAO RP
Source: Biochem Mol Biol Educ. 2020;1–7.
Synthesis and Biological Evaluation of 4/5-Aroyl-2-aminoimidazoles as Microbial Biofilm Inhibitors
Authors: Rasapalli S, Sammeta VR, Singh S, Golen JA, Semerdzhiev D, YANG B, Silby M, RAO R, Ali A, Schiffer CA, and Savinov SN
Source: ChemistrySelect 2020, 5, 5965 –5969
Coordinated host-pathogen transcriptional dynamics revealed using sorted subpopulations and single macrophages infected with Candida albicans
Authors: Munoz JF, DELOREY T, Ford C, Li BY, Thompsom DA, RAO RP, Cuomo CA
Source: Nat Commun, 10, 1607
Probiotic Yeasts Inhibit Virulence of Non-albicans Candida Species
Authors: Kunyeit, L., Kurrey, NK, Anu-Appaiah KA, & RAO, R. P
Source: mBio, 10, e02307-19.
The nematode Caenorhabditis elegans – A versatile in vivo model to study host-microbe interactions
Authors: ISSI, L., RIOUX, M, RAO R.
Source: J. Vis. Exp. 128(e56487).
Whole genome sequence of the heterozygous clinical isolate Candida krusei 81-B-5
Authors: Cuomo, C, Shea, T, YANG, B, RAO R, Forche, A
Source: Jul10, pii:G3.10.1534/g3.117.043547.
A pretherapeutic coating for medical devices that prevents the attachment of Candida albicans
Authors: VARGAS-BLANCO D, Lynn A, ROSCH J, NORELDIN R, SALERNI A, Lambert C, RAO R,
Source: BMC: Ann Clin Microbiol Antimicrob 16(1):41
Zinc Cluster Transcription Factors alter virulence in Candida albicans
Authors: ISSI, L., Farrar, R. A., PASTOR, K., LANDRY, B., DELOREY, T. M, Bell, G., Thompson, D. A., Cuomo, C. A., RAO, R. P.
Source: Genetics, February 2017 205: 559-576.
The evolution of drug resistance in clinical isolates of Candida albicans
Authors: Ford CB, Funt, JM, Abbey, D, ISSI, L., Oliver, BG, Guiducci, C, Martinez, DA, DELOREY, T, Li, BY, White, TC, Cuomo, C, RAO, R. P, Berman, J, Thompson, D, Regev, A.
Source: eLife;4:e00662.
Remarks: Featured in eLIFE insight (figure generated in RAO lab).
Chemical screening identifies a small molecule inhibitor of C
Authors: Fazly A, JAIN C, Dehner AC, ISSI L, Lilly E, Fidel PL, RAO R. P *, Kaufman PD* (* Corresponding authors)
Source: albicans adhesion, morphogenesis and pathogenesis. PNAS 110(33): 13594-13599.
The role of Candida albicans AP-1 protein against host derived ROS in in-vivo models of infection
Authors: JAIN C, PASTOR K, Gonzalez AY, Lorenz MC, RAO R. P.
Source: Virulence 4: 67-76.
Aberrant synthesis of Indole-3-acetic acid in Saccharomyces cerevisiae triggers morphogenic transition, a virulence trait of pathogenic fungi
Authors: RAO R. P, HUNTER A, KASHPUR O and Normanly J
Source: Genetics 185 (1): 211-220.
Remarks: Featured in the highlights section of the journal. Featured in Faculty of 1000.
A Patho-assay using Scerevisiae and C. elegans reveals novel roles for yeast AP-1, Yap1 and host dual oxidase BLI-3 in fungal pathogenesis
Authors: J CHARU, YUN M, Politz S. M, RAO R. P.
Source: Eukaryotic Cell. 8 (8) 1218-1227
Remarks: Featured in the Science highlights of the NECN Cable News network.
Expression Profiling of Auxin-Treated Arabidopsis Roots: Toward a molecular analysis of lateral root emergence
Authors: Laskowski M, Biller S, Stanley K, Kajstura T, PRUSTY R.
Source: Plant and Cell Physiology 47(6): 788-792.
SCH9, a putative kinase from Scerevisiae, affects HOT1-stimulated recombination
Authors: PRUSTY R, Keil RL.
Source: Molecular Genetics and Genomics, 272: 264-274.
The plant hormone, Indole acetic acid, induces invasive growth in Saccharomyces cerevisiae
Authors: PRUSTY R, Grisafi P, Fink GR.
Source: Proceedings of the National Academy of Sciences USA, 101(12): 4153-57.
Remarks: Featured in Faculty of 1000 ‘must read papers’
Ribosomal DNA replication fork barrier and HOT1 recombination hot spot: shared sequences but independent activities
Authors: Ward TR*, Huong MJ*, PRUSTY R*, Lau CK, Keil RL, Fangman WL, Brewer BJ. (* Equal contribution)
Source: Molecular and Cellular Biology, 20(13): 4948-57.
Elimination of yeast replication block protein Fob1p extends the life span of mother cells
Authors: Defossez P, PRUSTY R, Kaeberlein M, Lin S, Ferrigno P, Silver PA, Keil RL, Guarente L.
Source: Molecular Cell, 3: 447-455.
Remarks: Featured in News and Views in Nature Genetics, May 1999, 22: 4-6.
ENOD 8, a Medicago early nodulin gene, expressed in empty nodules
Authors: Dickstein R, PRUSTY R, Peng T, Ngo W and Smith ME
Source: Molecular Plant Microbe Interactions, 6: 715-721.
Group

| LAB MEMBERS | Undergraduate students |
| Post-doctoral Fellow
Lohith Kunyeit |
Susanna Oppong Alex Guerra Addison Writ Jones Michelle Pan David Datta Meredith Rioux Monet Norales Daira Daly Rose Awada Erica Friel Nina Murphy-Cook Natalie Fabrizio Brittney Lambert Meredith Rioux Jeffery Letourneau Heather Bartlett Rony Noreldin Jonah Rosch Chietara Japutra Allison Simpson Michael Boyd Damien Cabral Nouran Abdelfattah Giles Chikering Stephanie Post Tracy Sears Vicky Mason Danica Rili Margaret Chiasson Benjamin Landry Kurtis McCannell Kelly Pastor Christy Royer Nick Dufour Pamela Levandowsky Muxun Zhao Kyle Peet Linsley Kelly Brendan O’Brien Rachel Robillard Meryl Gray Zoe Lentz |
| Doctoral students
Bo Yang |
|
| Masters’ students
Sam Bryce |
Contact
Worcester Polytechnic Institute
100 Institute Rd.
Worcester, MA 01609 USA
Email: rpr@wpi.edu
Broad Institute
7 Cambridge Center
Cambridge, MA 02143 USA
Email: rpr@broadinstitute.org

