The Worcester Head Injury Model (WHIM)
|
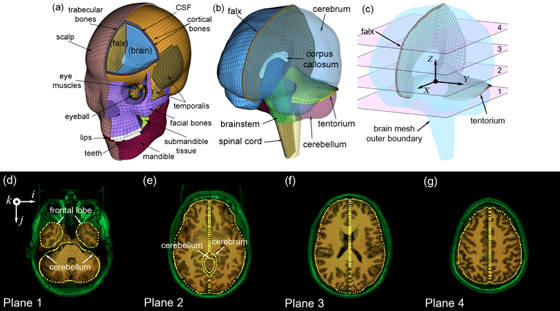
The biomechanical mechanisms behind traumatic brain injury (TBI) have been an active research focus for more than 70 years. However, the field is still largely focused on impact kinematics or estimated brain responses in generic regions from single head impact to predict a binary brain injury status on a population basis. A much more graded injury characterization, both spatially and temporally, is needed especially for mild TBI (mTBI) including sports concussion. There is a pressing need to study the distribution of injury from cumulative impacts, to correlate with brain functional alteration or recovery, and to investigate on an individual basis. In one line of research in this area, we are building pre-computed brain response atlases or large “lookup tables” to allow real-time strain and pressure feedback for a given impact. This exceptional efficiency may enable future studies to focus on region-specific, tissue-level mechanical responses, rather than relying solely on global kinematic variables or their variants that have plagued early and contemporary TBI research to-date. Separately, we are also pushing to integrate advanced neuroimaging into brain biomechanics modeling (e.g., incorporate whole-brain tractography; (Zhao et al., 2016)). This work builds on top of the Worcester Dartmouth Head Injury Model (WHIM; formerly known as the Dartmouth Head Injury Model, DHIM). This is a subject-specific finite element model of the human head created from high-resolution MRI of an actual athlete (Ji et al., 2015). It also allows the creation of other subject-specific head models via image registration. The WHIM has a high mesh quality and geometrical accuracy, based on quantitative measures. The model was successfully validated against relative brain-skull displacement and pressure responses from cadaveric impacts as well as strain responses from a live human volunteer, with performance rated as “good” to “excellent” according to an established fidelity rating (Ji et al., 2015, 2014; Zhao et al., 2015). |
| We are sharing with the research community our pre-computed brain strain response atlas (Ji and Zhao, 2015; Zhao and Ji, 2015b) as well as the brain pressure response atlas (Zhao and Ji, 2016) for research use and evaluation, along with the associated analysis software. We are open to any constructive suggestions, recommendations, and comments to improve our work further.
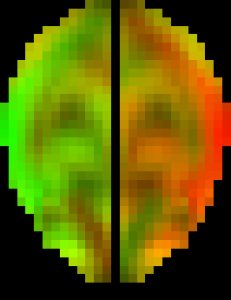
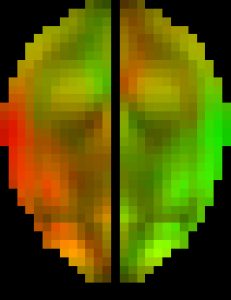
The original pcBRA-strain encodes the peak maximum principal strain at every element, which requires an explicit definition of the element structure. To facilitate the exchange of data and results, the pcBRA-strain was spatially resampled to generate a set of 3D grid data points encoded with strain values, analogous to stacked medical images such as MRI or CT. This allows direct application of many existing visualization and analysis routines, without the need for element definition. Here is the data (right click to save, may need to rename to “.mat” extension) and demo program in Matlab, which describes how to generate the following “happy” and “neutral” faces (they were created by overlaying two symmetric strain maps on an axial image plane in “occipital” and “frontal” impacts, respectively. The given subset of data only supports the “neutral” face on the right.
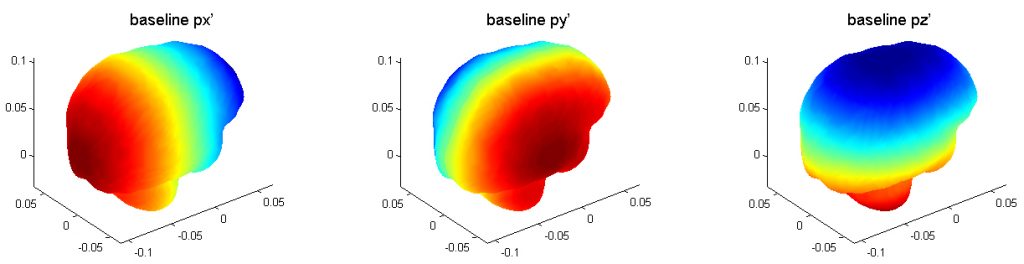
The three pre-computed pressure responses are shown below. They are linearly superimposed to obtain whole-brain pressure in an arbitrary translational head impact. Here is the data (right click to save, may need to rename to “.mat” extension) and demo program in Matlab.
References:
|
Permission to use for Research Purposes Only. All Other Rights Reserved.